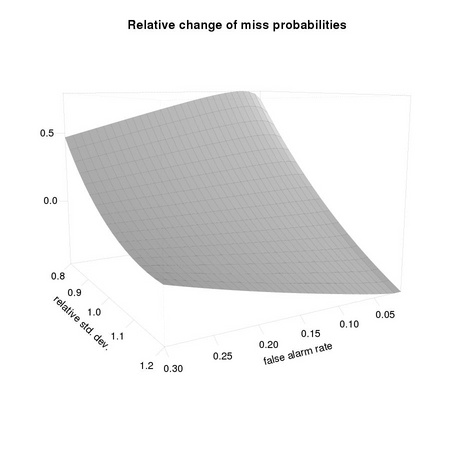
We want to visualize the impact of errors in the estimation of the covariance matrix on PD, the detection probability, at different operating conditions PF, as typical in the Neyman-Pearson paradigm. We refer to a single dimensional case, assuming that the distance of class means is 1: the difficulty of the problem is changed by changing the (common) standard deviation σ describing the two distributions. As easily checked from the results of Section TM3.3, we have that σ0 = σ-1. We want to compute the impact on PD(σ0) given that we set the operating condition using PF(σ′), where σ′ is our estimate of σ0 We first need to define a few functions, corresponding to Equation TM:3.15,
to Equation TM:3.57,
and to Equation TM:3.58,
By choosing σ = 1∕3, from which σ0 = 3, we get a reasonable testing case:
We then consider PF  [0.01, 0.3]
[0.01, 0.3]
and a moderately large range for σ′ = ασ0 α  [0.8, 1.2]:
[0.8, 1.2]:
We generate the sampling sequences:
14 pfs <- do.call(seq, as.list(pfRange))
15 pcs <- do.call(seq, as.list(pcRange)) * sigma0
and determine their lengths
from which we appropriately size the map:
We now hypothesize several estimated values σ′,
and for each of them, we build the function nu ν = ν(x) : PF(ν) = x:
We can now compute for a selected subset of operating conditions PF(σ′) based on our estimated standard deviation, the difference in the miss probability with respect to the correct one PD(σ0):